Command Palette
Search for a command to run...
Cambridge University Has Developed a Blood Cell Image Classifier; Its Diffusion Model Aids in Leukemia Detection, Surpassing the Capabilities of Clinical experts.

Blood cell imaging analysis plays a crucial role in clinical diagnosis and scientific research. The morphological characteristics of white blood cells, red blood cells, and platelets not only reflect the health of the blood system but may also reveal early disease signals, such as leukemia and myelodysplastic syndromes. However, traditional manual microscopic analysis relies on experienced experts for manual classification, which is inefficient, time-consuming, and subject to subjective bias.
In recent years, deep learning technology has been increasingly applied in the field of medical image analysis, and some studies have attempted to apply discriminative models, especially convolutional neural networks (CNNs), to blood cell morphology assessment. Although the best-performing discriminative ML classification models can approximate human performance in classifying cells into predefined categories, they primarily learn decision boundaries based on expert labels. Therefore,They are not naturally designed to capture a complete data distribution of cell morphology.This limitation reduces their capabilities, especially when faced with the inherent complexity and variability in clinical hematology data.
In this context,A research team from the University of Cambridge in the UK has proposed CytoDiffusion, a blood cell image classification method based on a diffusion model.It can faithfully model the morphological distribution of blood cells, achieve accurate classification, and has powerful abnormality detection capabilities, resistance to distribution shifts, interpretability, high data efficiency, and uncertainty quantification capabilities that surpass those of clinical experts.
The model outperforms state-of-the-art discriminant models in anomaly detection (AUC: 0.990 vs. 0.916), robustness to distribution shifts (accuracy: 0.854 vs. 0.738), and performance in low-data scenarios (balanced accuracy: 0.962 vs. 0.924).The comprehensive evaluation framework developed in this study establishes a multidimensional benchmark for hematological medical image analysis, which is expected to improve diagnostic accuracy in clinical settings.
The relevant research findings, titled "Deep generative classification of blood cell morphology," have been published in Nature.
Research highlights:
* Applying a latent diffusion model to blood cell image classification.
* We propose an evaluation framework that goes beyond standard metrics such as accuracy, incorporating robustness to distribution shifts, anomaly detection capabilities, and performance in low-data scenarios.
* Construct a new blood cell image dataset that includes imaging artifacts and annotator confidence, addressing key limitations of existing datasets.

Paper address:
https://www.nature.com/articles/s42256-025-01122-7
Follow the official account and reply to "CytoDiffusionGet the full PDF
Dataset address:
More AI frontier papers:
https://hyper.ai/papers
Datasets: Combining publicly available datasets and custom-built datasets
Data is the foundation of blood cell image analysis and a core guarantee for the performance and generalization ability of AI models. The CytoDiffusion team used five datasets, four of which are publicly available, and the other is a self-built dataset called CytoData.
The CytoData dataset is an anonymized dataset containing 2,904 blood smears from Addenbrooke's Hospital in Cambridge, totaling 559,808 single-cell images. Of these, 4,996 images are labeled with ten blood cell types, including erythroblasts, eosinophils, monocytes, and immature cells. The images were acquired using a CellaVision DM9600 system, and the annotations include confidence scores from each annotation expert, providing important reference for subsequent uncertainty quantification. CytoData also includes artifact categories to address common non-cellular structural interference in blood smears, which is of significant value in clinical applications.
Raabin-WBC, PBC, Bodzas, LISC as 4 public datasetsThe dataset encompasses blood cell images obtained using different microscopes, staining methods, and equipment. Raabin-WBC provides Test-A and Test-B partitions; Test-B uses different acquisition equipment than the training set to simulate domain shift. Due to the differences in equipment and staining methods, the LISC dataset places greater emphasis on the model's generalization ability.
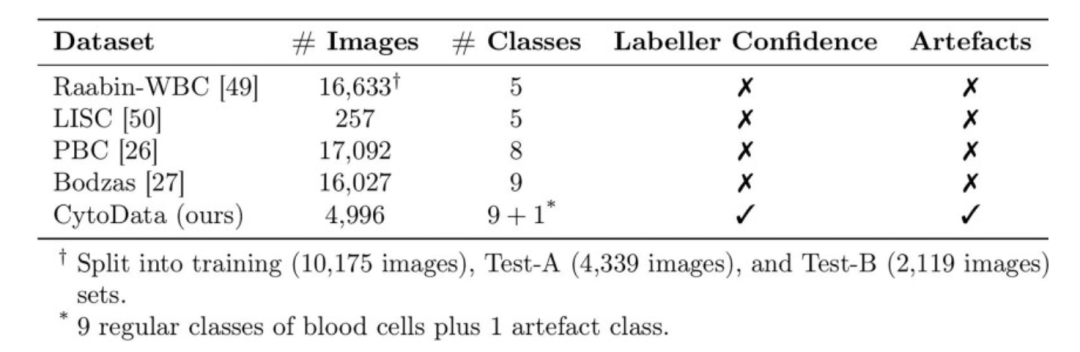
By combining these multi-source datasets, the team not only ensured the diversity of model training, but also provided a complete foundation for cross-domain performance evaluation, abnormal cell detection, and model testing under data-scarce conditions.
Dataset address:
Framework: Applying a diffusion model to blood cell image classification
CytoDiffusion’s core innovation lies in applying a diffusion model to blood cell image classification.Unlike traditional discriminative models, diffusion models have generative properties, enabling them to learn the complete distribution of an image and classify it through a noise prediction mechanism.
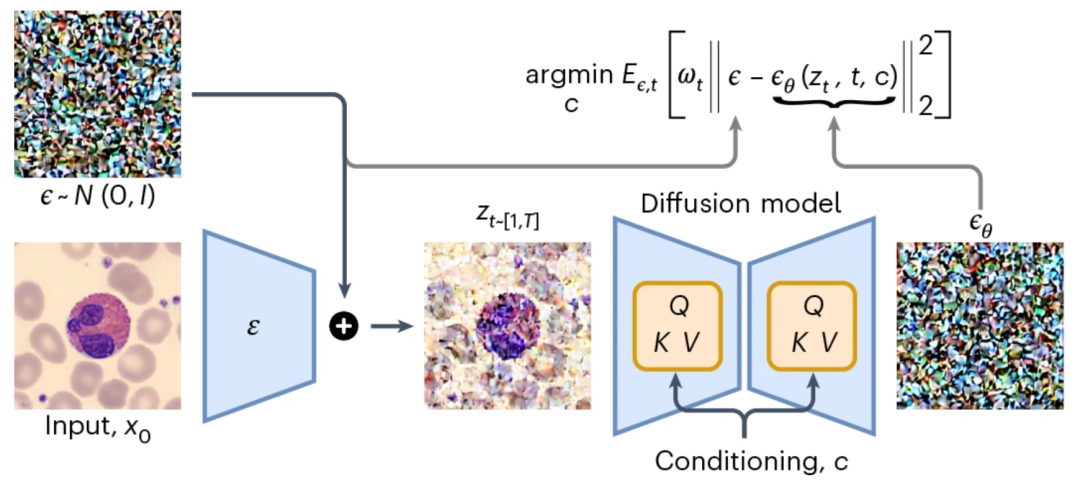
Model Principles
The core principle of the diffusion model is to define a forward diffusion process that transforms the data into a noise-like distribution by progressively adding noise. The model then learns an inverse process to denoise the data, thereby effectively reconstructing the original data distribution.
Latent space encoding:The input image is first mapped to the latent space by an encoder, and then Gaussian noise is added to form a noisy latent representation;
Conditional diffusion:The model generates noise predictions for each cell type and achieves classification by minimizing the error between the predicted noise and the actual noise.
Eliminate gradually:Iterative sampling is performed on all candidate categories, and impossible categories are eliminated step by step using the paired Student's t test until the final category is determined.
General training settings
The researchers used Stable Diffusion 1.5 as the base model.For category-based conditions, it bypasses the tokenizer and text encoder, directly providing one-hot encoded vectors for each category. These vectors are vertically copied and horizontally padded to match the expected 77 × 768 dimensional matrix. A batch size of 10, a learning rate of 10⁻⁵, and a linear warm-up of 1000 steps were used, with training on an A100-80GB GPU.
Training and reasoning
Researchers applied a variety of data augmentation methods during training, including random diagonal flips, random rotations (uniform sampling between 0 and 359 degrees), color jitter (brightness = 0.25, contrast = 0.25, saturation = 0.25, hue = 0.125), Mixup (α = 0.3, applied to conditional inputs rather than the target), and RandAugment (using default parameters).
Training used the AdamW optimizer (β1 = 0.9, β2 = 0.999, ϵ = 10⁻⁸, weight decay 0.01), mixed precision training (fp16), and exponential moving average (0.9999). All images were uniformly resized to 360 × 360 pixels.
During the inference phase, the same data augmentation methods as in the training phase were applied, but Mixup was excluded. Taking advantage of the fact that white blood cells are typically located at the image center, and to reduce interference from augmentation of image edge regions, inference error was calculated only within a 20-pixel radius of the image center in the latent space. The latent space dimension was 45 × 45 × 4.
Results Showcase: CytoDiffusion can help solve key challenges in clinical deployment.
Image generation and authenticity verification
The clinical application of artificial intelligence systems not only requires high performance but also demands that the models possess reliable representational capabilities. To demonstrate that CytoDiffusion learns the true morphological distribution of blood cells, rather than relying on "shortcuts" such as artifacts, researchers conducted a realism test.
Based on 32,619 training images, the blood cell images generated by CytoDiffusion are virtually indistinguishable from real images. Ten hematologists conducted identification tests on 2,880 images, achieving an overall accuracy of 0.523 (random guess level), sensitivity of 0.558, and specificity of 0.489. This performance is close to random guessing, indicating that even for experienced professionals, the blood cell images generated by CytoDiffusion are virtually indistinguishable from real images.
The ability to generate images that are almost indistinguishable from real images indicates that CytoDiffusion has successfully learned the true distribution of blood cell morphology, as shown in the figure below:
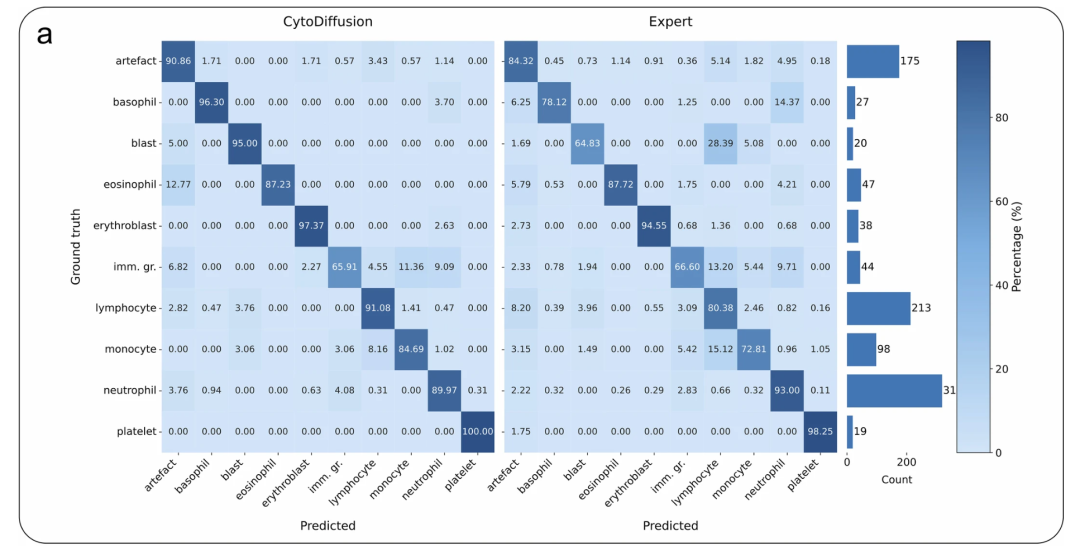
CytoData Comparison: The left matrix shows the results of CytoDiffusion, and the right matrix shows the average performance of human experts.
Classification performance
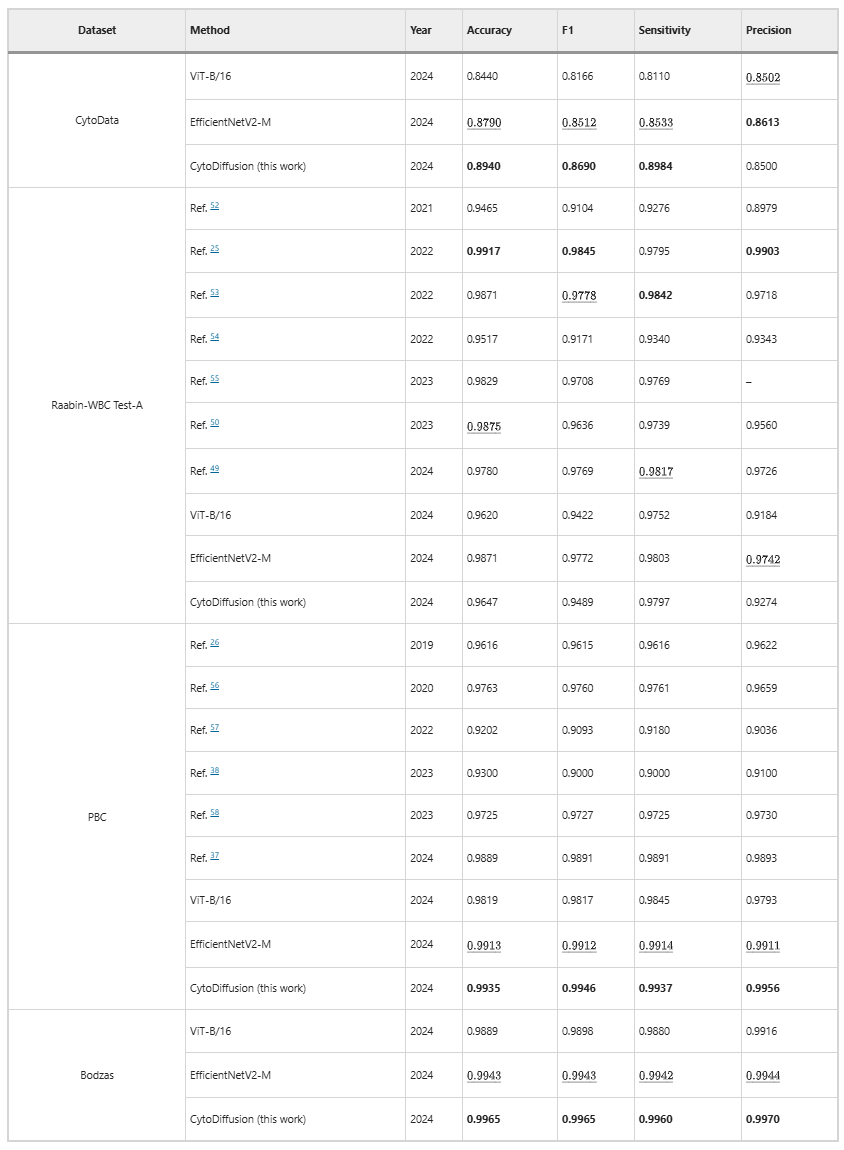
On four datasets (CytoData, Raabin-WBC, PBC, and Bodzas), CytoDiffusion achieves or surpasses the performance of traditional discriminative models. Particularly on CytoData, PBC, and Bodzas, the model achieves state-of-the-art performance, demonstrating that diffusion-based methods can match or outperform traditional discriminative models, as shown in the table below:
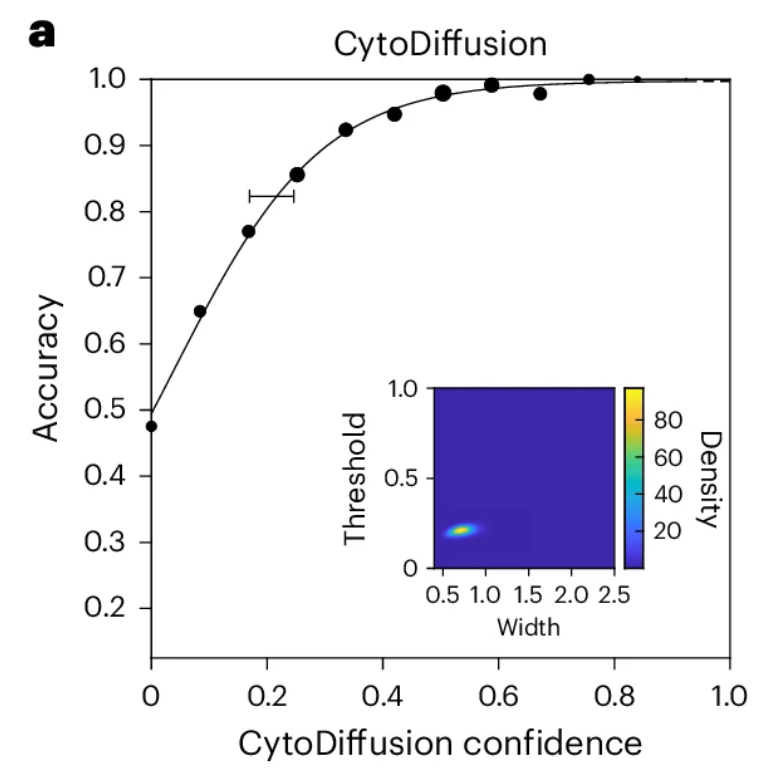
Uncertainty quantification is superior to that of human experts
Biological systems inherently possess irreducible uncertainty. In any analytical task, the measurement should not only focus on the accuracy of predictions but also on the uncertainty of the actor (whether human or machine).
Researchers used established Bayesian psychometric modeling techniques to derive the psychometric function of CytoDiffusion, as shown in the figure below.The results show that the model fits very well, and the posterior distributions of the key threshold and width parameters are very compact (embedded coordinate axes in the figure below).Although it cannot be directly measured, these results suggest that the uncertainty of CytoDiffusion is primarily dominated by a chance component, and its behavior closely resembles that of an ideal observer.
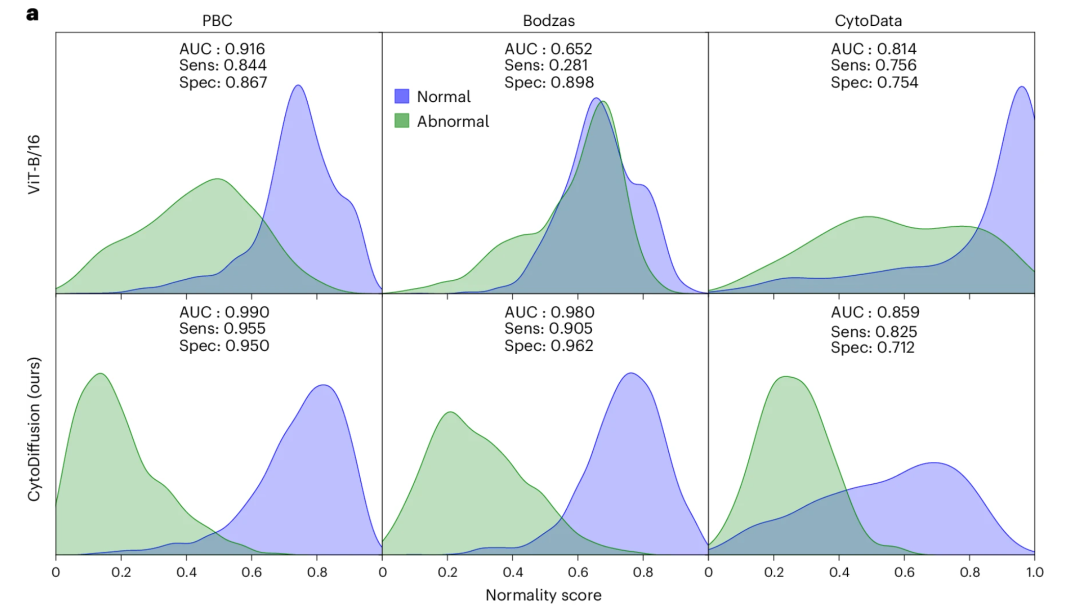
Abnormal cell detection and performance in low-data environments
For abnormal cell detection, CytoDiffusion achieved high sensitivity (0.905) and high specificity (0.962) when using primitive cells as the abnormality category in the Bodzas dataset. In contrast, ViT showed extremely low sensitivity (0.281), clearly failing to meet the needs of clinical applications, as shown in the figure below:
In low-data environments, with only 10–50 training images per class, CytoDiffusion significantly outperforms EfficientNetV2-M and ViT-B/16, demonstrating its efficient learning capability under data-scarce conditions, as shown in the figure below:
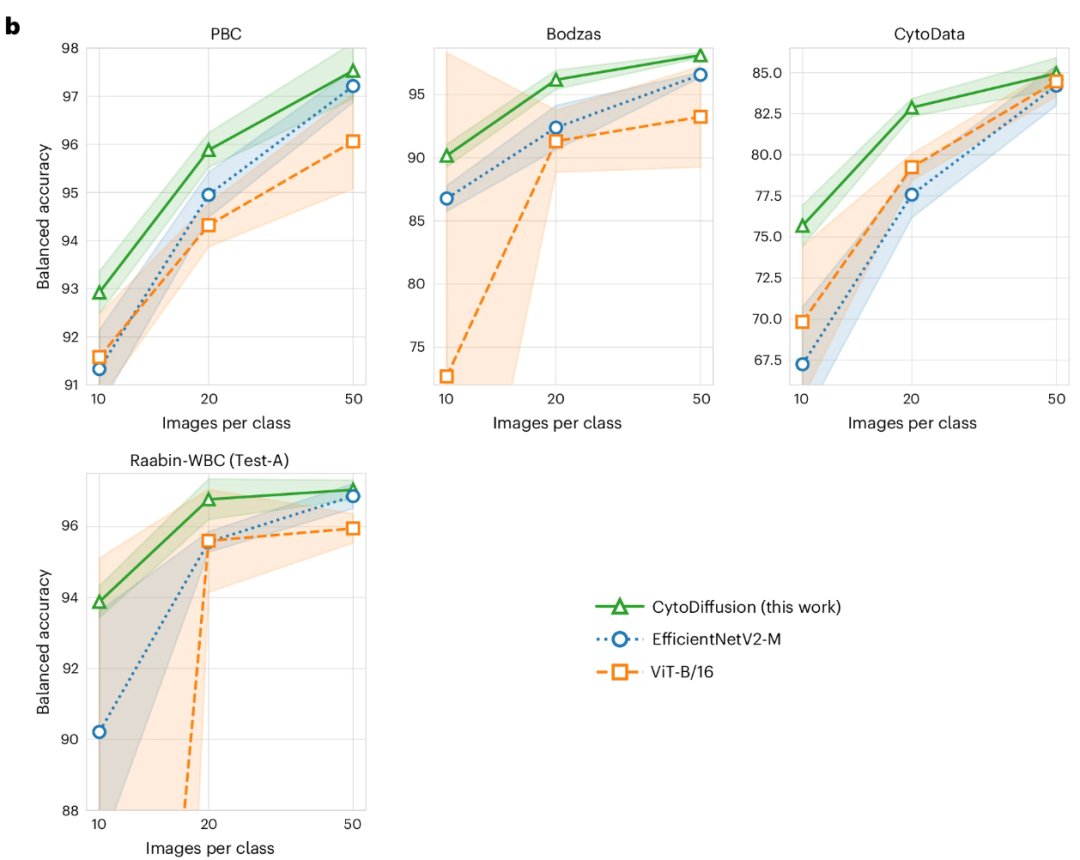
Model generalization ability
To evaluate the model's generalization ability, researchers tested its performance on different data domains. Models trained on Raabin-WBC were tested on the Test-B (using different microscopes and cameras) and LISC (different microscopes, cameras, and staining methods) datasets; models trained on CytoData were tested on PBC and Bodzas. CytoDiffusion achieved state-of-the-art accuracy on all four datasets.This consistency advantage across varying degrees of domain drift demonstrates that CytoDiffusion is robust to dataset variations and possesses good generalization ability in real-world clinical scenarios.
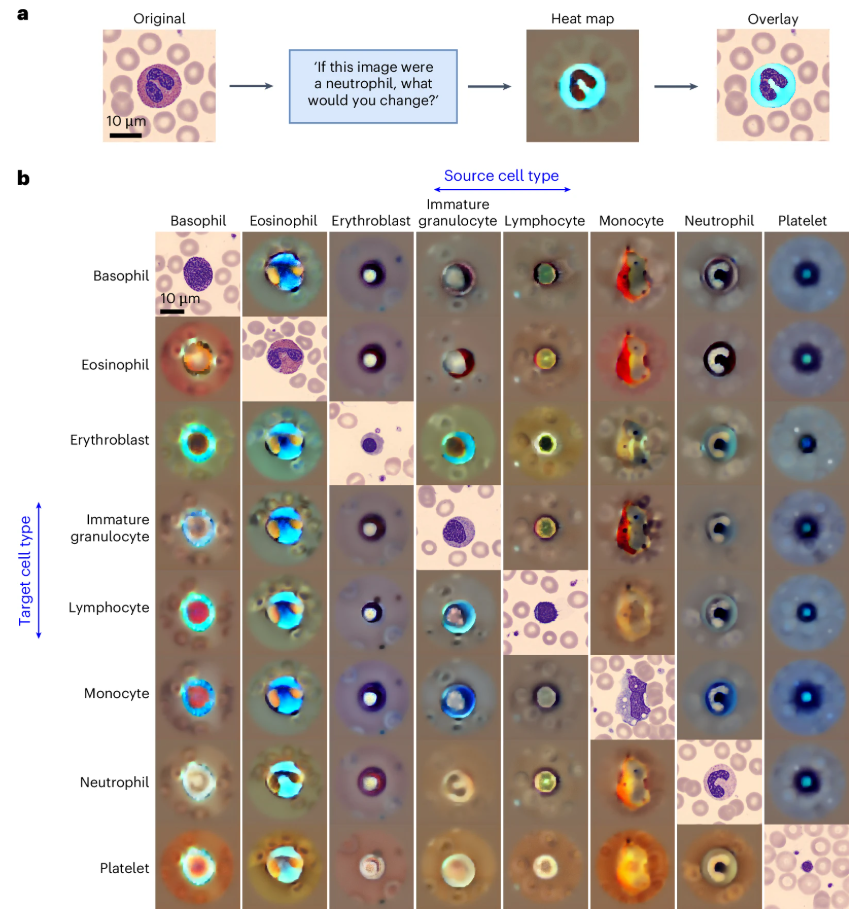
Explainability verification
Through counterfactual heatmap analysis, the model can identify key morphological features within cells, as shown in the figure below. For example, in the transition from monocytes to immature granulocytes, the model highlights differences in cytoplasmic acidity and vacuolar filling characteristics. This visualization not only validates the model's learning ability but can also be used to detect potential biases, ensuring that classification criteria align with clinical scientific logic.
Diffusion models are proving groundbreaking in the biomedical field.
CytoDiffusion's research not only demonstrates the potential of diffusion models in blood cell morphology classification, but also reflects the rapid rise of diffusion-based generative frameworks in the entire biomedical field, showcasing groundbreaking value in multiple application scenarios.
For example, medical data is often limited and faces significant privacy concerns, making data acquisition and annotation a major challenge. Diffusion models can address this issue by generating synthetic medical images.This helps train deep learning models and improves the accuracy of medical image analysis;In addition to generating ordinary medical images, expanding...The dispersion model can also be used to generate medical images for specific conditions (such as tumors, fractures, etc.).This is especially important when training medical diagnostic models, as it can provide more training samples for rare diseases or images that are difficult to obtain. At the same time, diffusion models can generate high-quality, clear and realistic images, which not only helps improve the diagnostic accuracy of doctors, but also helps assist medical AI systems in making more accurate predictions.
In many clinical and research scenarios, the scarcity of high-quality medical image datasets hinders the potential of artificial intelligence (AI) in clinical applications. In December 2024, Professor Kang Zhang and Professor Jia Qu from the Eye Hospital Affiliated to Wenzhou Medical University, and Researcher Jinzhuo Wang from Peking University were the corresponding authors, while Dr. Kai Wang from Peking University and Dr. Yunfang Yu from Sun Yat-sen Memorial Hospital of Sun Yat-sen University were co-first authors.A novel framework, MINIM, based on a diffusion model, was developed. This model can generate medical images of various imaging modalities of different organs based on text commands.Clinician assessments and rigorous objective measurements validated the high quality of MINIM-generated images. When faced with unprecedented data domains, MINIM demonstrated enhanced generative capabilities, showcasing its potential as a general medical AI (GMAI).
Paper Title:Self-improving generative foundation model for synthetic medical image generation and clinical applications
Paper address:https://www.nature.com/articles/s41591-024-03359-y
In cell biology research, living cells are complex dissipative systems far removed from chemical equilibrium. How their collective response to external stimuli has always been a core scientific question that scientists are striving to uncover. November 2025,Research teams from Columbia University, Stanford University, and other institutions have developed the Squidiff computational framework.This framework, built upon a conditionally denoised diffusion implicit model, can predict transcriptomic responses in different cell types under differentiation induction, gene perturbation, and drug treatment. Its core advantage lies in its ability to integrate explicit information from gene editing tools and drug compounds: in predicting stem cell differentiation, it can not only accurately capture transient cell states but also identify non-additive gene perturbation effects and cell-specific response characteristics. The research team further applied Squidiff to vascular organoid research, successfully predicting the effects of radiation exposure on various cell types and evaluating the protective efficacy of radiation-protective drugs.
Paper Title:Squidiff: predicting cellular development and responses to perturbations using a diffusion model
Paper address:https://www.nature.com/articles/s41592-025-02877-y
It is foreseeable that as generative basic models mature further in the medical field, diffusion models will be implemented in more real clinical scenarios, becoming an important foundation for general medical intelligence, bringing higher reliability, stronger generalization ability and wider application space for future medical image diagnosis, disease prediction and intelligent decision-making.
References:
1.https://www.nature.com/articles/s42256-025-01122-7
2.https://www.nature.com/articles/s41592-025-02877-y
3.https://mp.weixin.qq.com/s/9JEt-QwFxngv9XC0hSIcnw
4.https://bbs.huaweicloud.com/blogs/448218