Command Palette
Search for a command to run...
MedXIAOHE: A Comprehensive Recipe for Building Medical MLLMs
MedXIAOHE: A Comprehensive Recipe for Building Medical MLLMs
Abstract
We present MedXIAOHE, a medical vision-language foundation model designed to advance general-purpose medical understanding and reasoning in real-world clinical applications. MedXIAOHE achieves state-of-the-art performance across diverse medical benchmarks and surpasses leading closed-source multimodal systems on multiple capabilities. To achieve this, we propose an entity-aware continual pretraining framework that organizes heterogeneous medical corpora to broaden knowledge coverage and reduce long-tail gaps (e.g., rare diseases). For medical expert-level reasoning and interaction, MedXIAOHE incorporates diverse medical reasoning patterns via reinforcement learning and tool-augmented agentic training, enabling multi-step diagnostic reasoning with verifiable decision traces. To improve reliability in real-world use, MedXIAOHE integrates user-preference rubrics, evidence-grounded reasoning, and low-hallucination long-form report generation, with improved adherence to medical instructions. We release this report to document our practical design choices, scaling insights, and evaluation framework, hoping to inspire further research.
One-sentence Summary
ByteDance XiaoHe Medical AI presents MedXIAOHE, a medical vision-language model using entity-aware continual pretraining and tool-augmented agentic training to enhance reasoning and reduce hallucinations, outperforming closed-source systems in real-world clinical diagnostics with verifiable, evidence-grounded outputs.
Key Contributions
- MedXIAOHE introduces an entity-aware continual pretraining framework that structures heterogeneous medical data using a medical entity taxonomy, improving coverage of rare diseases and reducing long-tail knowledge gaps across modalities.
- The model enables expert-level diagnostic reasoning through reinforcement learning and tool-augmented agentic training, producing multi-step, verifiable decision traces that align with clinical workflows.
- Evaluated across 30+ public and in-house benchmarks, MedXIAOHE achieves state-of-the-art performance and demonstrates improved reliability via evidence-grounded generation, low hallucination rates, and adherence to medical instruction rubrics.
Introduction
The authors leverage a medical vision-language foundation model called MedXIAOHE to address the gap between benchmark performance and real-world clinical usability. Prior medical VLMs often struggle with long-tail conditions, hallucination in report generation, and inconsistent evaluation protocols—limiting their reliability in heterogeneous, high-stakes clinical environments. MedXIAOHE introduces an entity-aware continual pretraining framework that organizes diverse medical data to broaden knowledge coverage, paired with reinforcement learning and tool-augmented agentic training to enable verifiable, multi-step diagnostic reasoning. It also integrates evidence-grounded generation and a unified benchmark suite with standardized protocols to improve reproducibility and clinically relevant evaluation.
Dataset
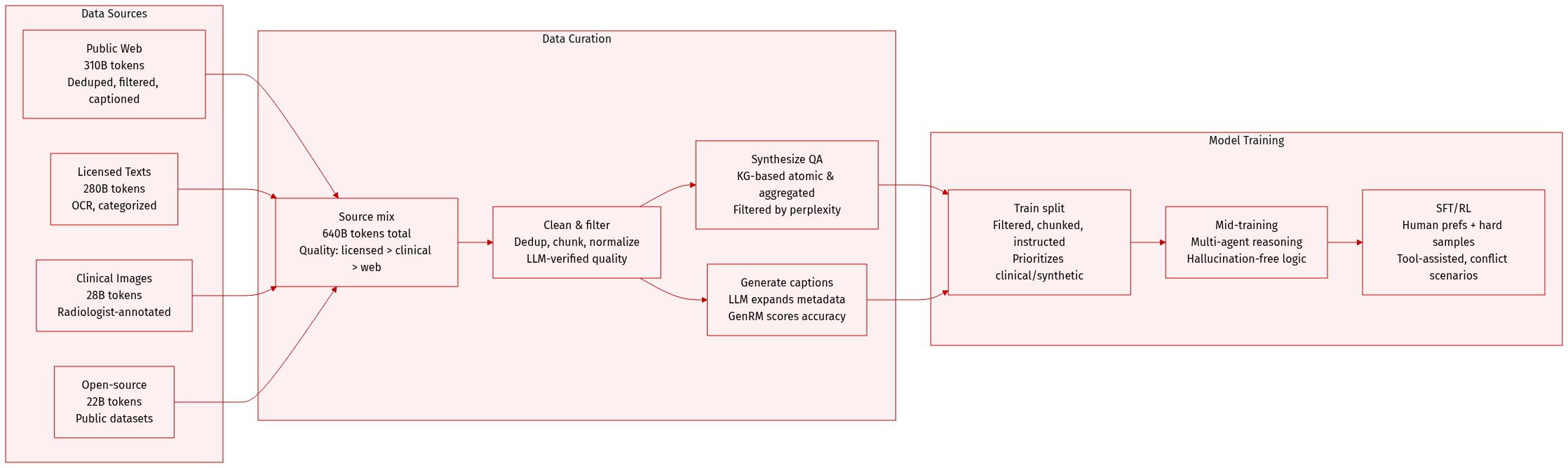
The authors use a meticulously curated, multimodal medical pretraining corpus totaling ~640B tokens, sourced from public web data, licensed medical texts, clinical images, and open-source datasets. Here’s how they build and use it:
-
Dataset Composition & Sources
- Public web: 310B tokens, enhanced with topic classification and updated captions.
- Licensed books/papers: 280B tokens, extracted via OCR with emphasis on recognition accuracy.
- Clinical lesion images: 28B tokens.
- Open-source datasets: 22B tokens.
- Total: ~640B tokens, with quality inversely proportional to volume (licensed > clinical > web).
-
Key Subset Details
- Public Web: Lightweight deduplication → model-based quality filtering → text normalization → image-text relevance checks.
- Licensed Texts: OCR applied to scanned materials; fine-grained categorization prioritized.
- Clinical Images: High-resolution, expert-annotated by radiologists (lesion localization, progression, differential notes).
- Synthetic QA: Built from a medical knowledge graph — Atomic QA (node/edge facts) and Aggregated QA (multi-fact synthesis via LLMs). Filtered for self-containedness and low perplexity.
- OCR Images: 9M medical report images selected via weakly supervised ViT-Base classifier; distilled via Seed1.5-VL; augmented with 10% rotation.
- Grounding Data: Generated via internal captioning + MLLM verifier to extract only visually grounded entities.
- Human-in-the-loop: Physicians annotate images (anatomy, lesions, diseases), then LLMs synthesize VQA pairs; filtered via general VLM (relevance/plausibility) and expert VLM rollouts for medical correctness.
-
Data Usage in Training
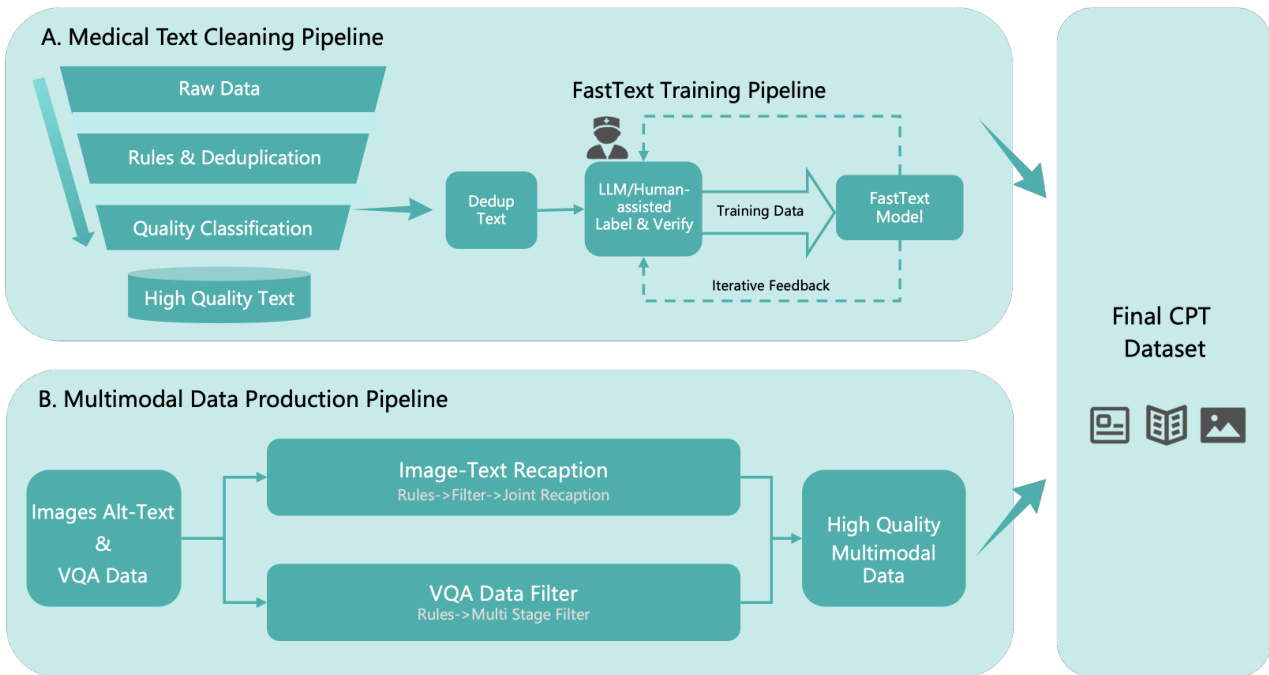
- Training split: Cleaned via 3-stage pipeline — global deduplication (hash-based), rule-based chunking (sliding window), model-based filtering (self-bootstrapped medical FastText classifier refined via LLM verification).
- Mixture ratios: Not explicitly stated, but synthetic QA and clinical image data are prioritized to reduce web noise.
- Processing: Text normalized, long docs split, captions rewritten via LLM prompts, instruction pairs generated at multiple granularities (descriptive, diagnostic, localization, reasoning).
- Caption quality: Filtered via GenRM trained on human-annotated rubrics (anatomical accuracy, morphological precision, evidence grounding).
-
Cropping & Metadata
- Text: Chunked via sliding window for manageable input lengths.
- Images: Metadata extracted from DICOM headers or structured reports (anatomy, finding, severity, laterality); converted to JSON → LLM-expanded into narrative captions.
- Grounding: Visual regions linked to text via automated entity extraction verified against image content.
- OCR: Real-world distortions preserved (blur, glare, occlusion) for robustness; filtered for noise via rule-based cleanup.
-
Mid-Training & SFT/RL Data
- Mid-training: Synthesizes reasoning trajectories via multi-agent consensus and chain-of-thought pipelines; targets hallucination-free, clinically grounded logic.
- SFT: Human preferences derived from multi-VLM consensus; synthetic data augmented with prompt rewriting and long-tail oversampling.
- RL: Prioritizes hard samples (60–80% SFT accuracy); adds tool-assisted and conflict scenarios; uses atomic evaluation points for clear reward signals.
-
Evaluation Benchmarks
- Public: Six categories — Visual Diagnosis, Medical Imaging, Diagnosis, Medical Text, Report Generation, Instruction Following — with multilingual and multimodal coverage.
- Inhouse: Real-world VQA (100K+ clinical questions), OCR (distorted patient-captured reports), Caption (lesion-focused, physician-verified key points scored via reward/penalty rubric).
Method
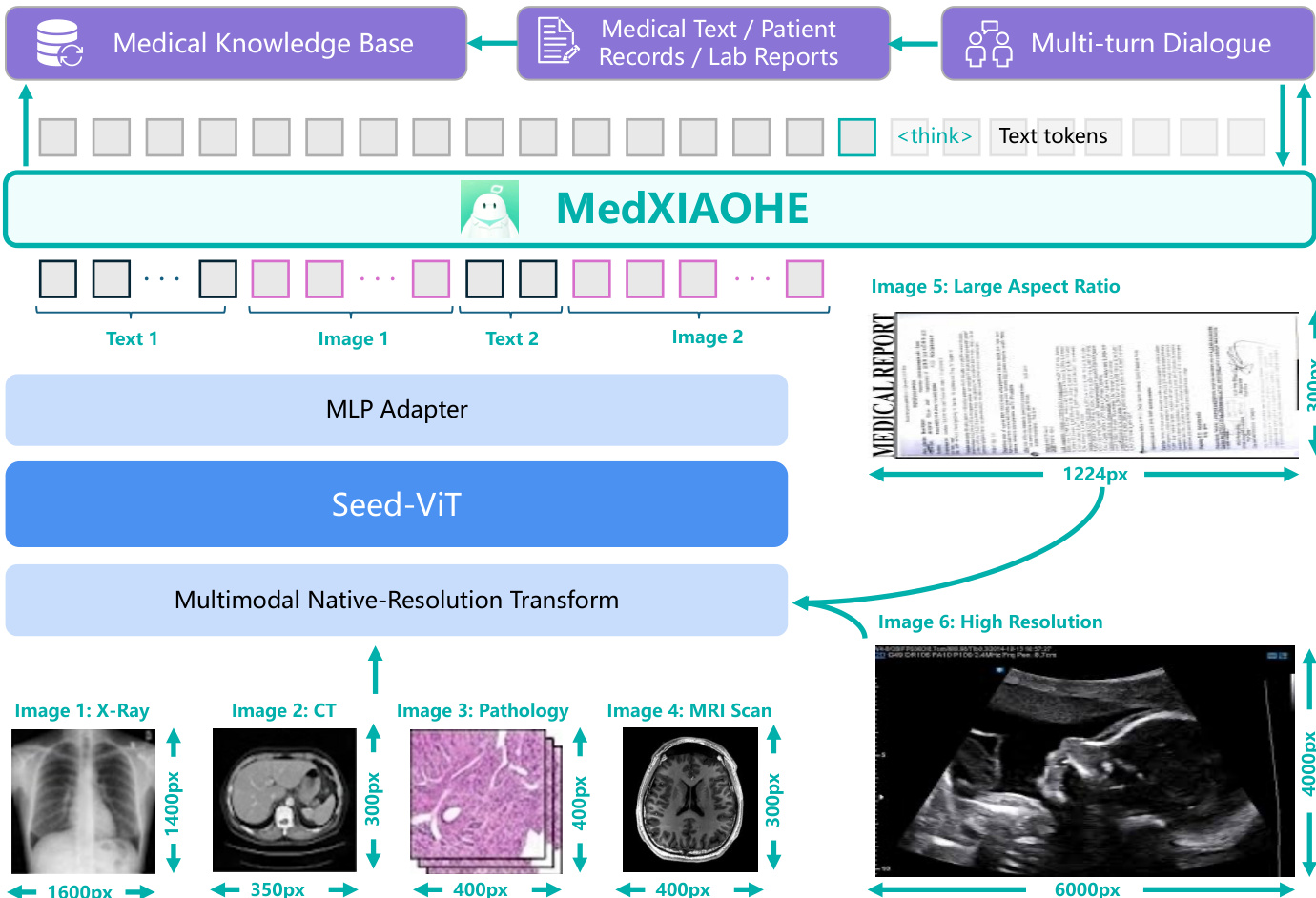
The authors leverage a unified multimodal decoder architecture for MedXIAOHE, built upon the Seed vision-language foundation model. The core design integrates a powerful vision encoder with a large language model (LLM), enabling seamless processing of interleaved visual and textual inputs. As shown in the framework diagram, the system accepts diverse modalities—including medical text, patient records, lab reports, and multi-turn dialogue—alongside high-resolution clinical images such as X-rays, CT scans, pathology slides, and MRI scans. These inputs are processed through a multimodal native-resolution transform that preserves spatial fidelity, followed by a Seed-ViT backbone that encodes images into visual tokens. A lightweight MLP adapter then maps these visual features into the LLM’s embedding space, allowing the autoregressive decoder to perform instruction following, reasoning, and generation conditioned on the full context.
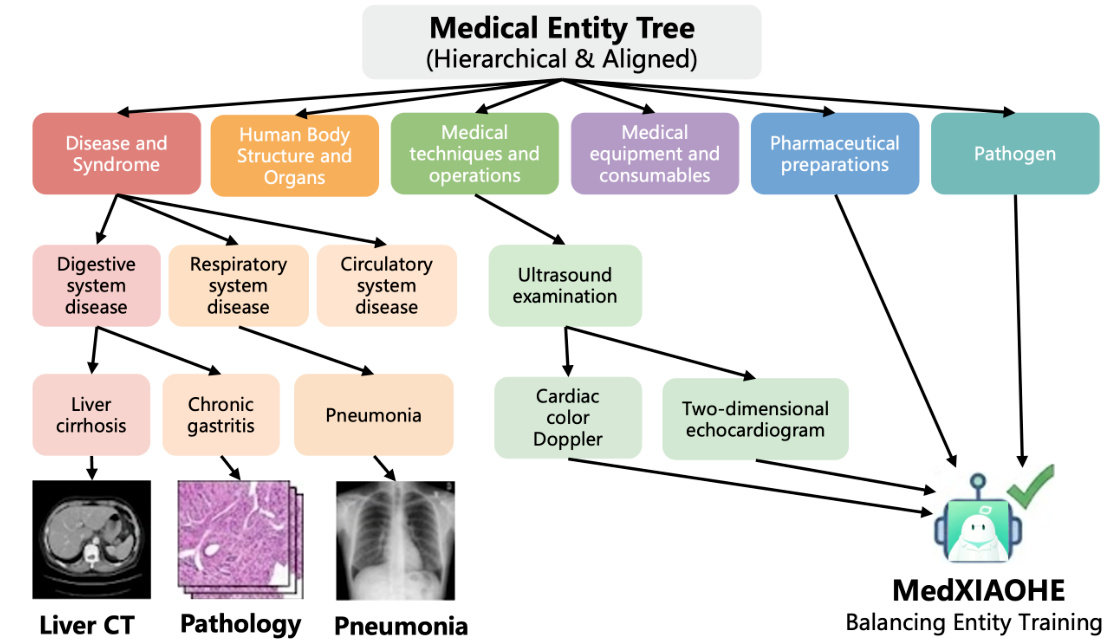
To support domain-specific robustness, the authors construct a Medical Entity Tree (MET), a hierarchical taxonomy comprising 1.4 million entities organized into five tiers. This structure serves to balance entity training, quantify knowledge coverage, and guide targeted data acquisition. The MET is built through a three-stage pipeline: initial high-efficiency entity extraction via LLMs with strict JSON formatting; joint typing and hierarchical clustering using K-Means and frequency-weighted aggregation; and controlled expansion via incremental tree attachment. During expansion, a ReAct Agent resolves classification conflicts by retrieving external evidence and applying domain-specific principles such as etiological dominance and specificity, ensuring auditability and dynamic adaptation to emerging medical knowledge.
The training process employs a single-stage continual pre-training (CPT) recipe that replaces random shuffling with a curriculum-driven data ordering strategy. The authors first train a lightweight warm-up model to generate fixed-length embeddings for each sample, then apply UMAP and HDBSCAN to cluster semantically coherent examples. Clusters are ranked by compactness score—lower scores indicating higher intra-cluster similarity—and ordered to form an easy-to-hard curriculum. Smooth transitions between clusters are enforced via mixed regions and replay buffers to mitigate forgetting and gradient conflict. This structured exposure enables stable joint optimization of medical specialization and general multimodal reasoning without explicit stage-wise freezing.
For reasoning capability development, the authors synthesize high-quality medical QA data using multiple pipelines. KG-guided QA synthesis generates multi-hop questions by sampling complex paths from a medical knowledge graph, masking intermediate attributes to increase difficulty. Multi-expert reject sampling employs ensemble models to produce diverse reasoning trajectories, validated through dual-quality gates for outcome and logical soundness. Structured CoT synthesis enforces a four-stage clinical workflow (Understanding, Observation, Reasoning, Conclusion) and validates outputs across six dimensions to ensure hindsight-free, authentic thinking. Personalized Visual CoT introduces a dual-track paradigm that separates logical core construction from stylistic adaptation, preserving visual fidelity while enhancing reasoning depth.
To enhance agentic reasoning, the authors equip the model with a toolset including General Search, Medical Search (Drug Labels, Clinical Records), and Image Edit (Zoom In, Rotate). The training dataset is synthesized through a difficulty-filtered pipeline that generates multi-hop medical questions requiring tool use. A two-stage filtering process selects cases demanding localized perception and tool-augmented inference, ensuring the model learns to interact with external tools for complex diagnostic tasks. The training loop integrates these tool-augmented trajectories to build a reasoner that grounds conclusions in visual evidence and verifies details via external sources.

Post-training employs supervised fine-tuning (SFT) with instruction-following data synthesized from atomic instruction sets and graph-structured representations to model implicit reasoning. A reverse construction strategy infers compatible instructions from high-quality responses to improve constraint adherence. Reinforcement learning follows, guided by a multi-layered hybrid reward system that routes samples to rule-based or rubric-based graders. The rubric system combines dynamic contextual criteria with static domain standards, anchored to gold-reference responses. Process-supervised reasoning verification evaluates intermediate CoT blocks for framework completeness, logical robustness, and exploratory depth. Reward signals are fused non-linearly with a safety gating mechanism to ensure clinical accuracy, logical coherence, and regulatory compliance.
Finally, the authors implement an RFT-enhanced iterative curriculum strategy to reconcile heterogeneous data and layered rewards. Each iteration cycles through RFT distillation, foundation RL, specialization RL, and alignment RL phases. Dynamic hint-based curriculum scaffolds early learning, while entropy-aware adaptive regulation prevents exploration stagnation in later stages. This iterative approach enables stepwise capability integration without gradient interference or entropy collapse, producing a clinically reliable system capable of complex, tool-augmented reasoning across multimodal medical tasks.
Experiment
- Validated medical taxonomy comprehensiveness via semantic coverage metrics, showing near-complete forward coverage (>0.95) across clinical, corpus, and knowledge graph benchmarks, and revealing significant backward coverage gaps that confirm superior granularity in the constructed tree.
- Demonstrated top performance in Cure-Bench@NeurIPS 2025 for medical reasoning tasks, with models using tool-assisted analysis (e.g., ZOOM) to magnify and interpret imaging features, enabling accurate diagnoses such as identifying lung cancer from CT scans.
- Showed structured, grounded reasoning capabilities in radiology, including systematic X-ray analysis with anatomical segmentation and bounding box annotation, leading to precise findings like cardiomegaly and vascular congestion.
- Successfully linked clinical presentations (e.g., pancytopenia, splenomegaly) to peripheral smear morphology (hairy cells) to diagnose Hairy Cell Leukemia, correctly identifying TRAP positivity as the key marker.
- Achieved accurate extraction of lab values from deformed clinical reports using tool-assisted zooming and region-specific analysis, confirming out-of-range measurements like low MCH.
- Evaluated across multiple public medical benchmarks covering visual diagnosis, imaging, text, reporting, and instruction following, demonstrating broad competency in diverse medical AI tasks.
The authors use a semantic coverage metric to compare their Medical Entity Tree against established medical datasets, finding that their taxonomy captures over 95% of concepts from each target set. The lower backward coverage scores indicate their tree includes many additional long-tail and fine-grained entities not present in the baselines, supporting its broader scope. Results show the taxonomy is both highly inclusive of existing knowledge and more comprehensive than current benchmarks.

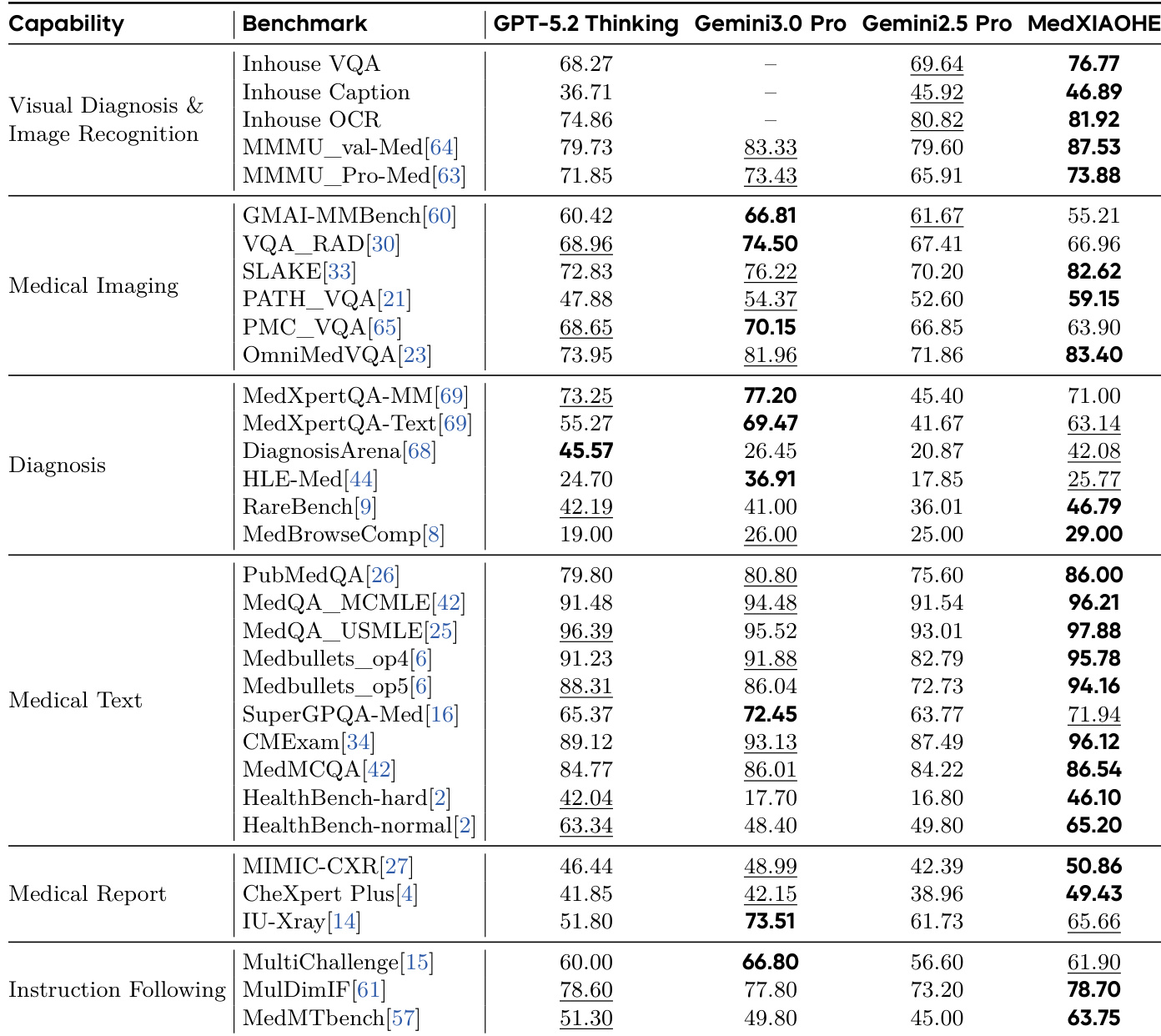
The authors evaluate their MedXIAOHE model across multiple medical benchmarks, comparing it against GPT-5.2 Thinking, Gemini3.0 Pro, and Gemini2.5 Pro. Results show MedXIAOHE consistently outperforms or matches leading models in visual diagnosis, medical imaging, diagnosis, medical text, and instruction following tasks, particularly excelling in specialized medical benchmarks like MedXpertQA-MM and MedMCQA. This indicates the model’s strong generalization and domain-specific proficiency in handling complex medical reasoning and multimodal tasks.